Genome Browser
The following link will give you access to the E. scolopes Genome Browser, which is being hosted by the University of Vienna.
http://metazoa.csb.univie.ac.at:8000/euprymna/jbrowse
Genome data (Belcaid et al., 2019):
http://metazoa.csb.univie.ac.at/data/v1/
Chromosomal-scale assembly and data (Schmidbauer et al., in review):
http://metazoa.csb.univie.ac.at/data/v2/
How to use this database
How the database works
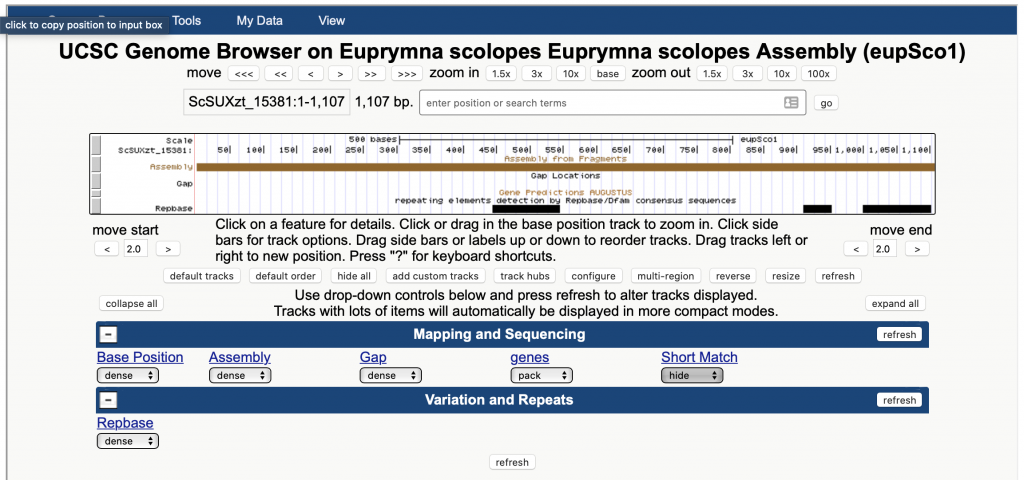
CephRes-gdatabase is made by a “genome browser” based on Kent et al. (2002) including some tools such as the table browser, BLAT and custom tracks. The features available allow also to connect with other databases such as Ensembl, Primer3Plus, etc.
This genome database browsing and mining initiative is a joint venture of CephRes and UniVienna.
Features implemented
Table browser:
The table browser enables the user to mine and filter big datasets using several parameters which produces subsets that can be compared.
BLAT
This Blast-Like-Alignment tool allows for fast alignments of any sequence with a minimum length of at least 25 bp.
Tools –Blat
Enter your query as a nucleotide or protein sequence
Output: List of best matches to your query.
Details: nucleotide sequence;scaffold id,matching regions;….
Browser: graphical view of your alignment on the genome
Custom tracks
It is possible to upload your private data to the genome browser:
My data – Custom tracks – Paste URLs or data
Enter the gene position: ScaffoldID_StartPos_StopPos_GeneID
or
Enter the gene sequence
My data – Custom tracks – Optional track documentation:
Enter your personal gene annotation.
More information on how to add custom tracks can be find here:
https://genome-euro.ucsc.edu/goldenPath/help/customTrack.html
In silico PCR By entering possible primers it is possible to simulate a PCR in silico against the entire genome, which will permit more efficient and specific primer design.
Sessions tool By creating a session you can save the current mode of the genome browser and go back to the same settings at a later time. For more info see: http://metazoa.csb.univie.ac.at/ugb/public/goldenPath/help/hgSessionHelp.html